Background
Tuberculosis (TB) is a deadly infectious disease, primarily affecting the lungs. It is caused by the pathogenic bacterium Mycobacterium tuberculosis (MTB). Transmission occurs when a person with active TB disease coughs and aerosolizes the bacteria, which then spread to other individuals. Symptoms of active TB include chest pain, prolonged cough, and blood in the sputum. Worldwide, approximately 1.4 million people die from TB each year, with most of the new cases and deaths occurring in developing countries.
Because TB is caused by a bacterium, it can often be treated using antibiotics, although the course of treatment typically extends 6 months or longer. Alarmingly, there is an increasing prevalence of TB strains that are resistant to the antibiotics typically prescribed, including multi-drug resistant (MDR) and extensively drug resistant (XDR) strains. MDR-TB infections are resistant to two of the first line TB antibiotics, isoniazid and rifampin, and XDR-TB infections are resistant to four types of antibiotics.
M. tuberculosis colonies are very slow-growing, so the recommended treatment of a new case of TB is 6 months of a combination of 2-4 different types of antibiotics. For MDR-TB, treatment with at least 4 effective antibiotics should last from 18 to 24 months. It is important that antibiotics with a strong likelihood of success be prescribed as quickly as possible and taken as directed for the entire prescribed length of time. Failure to do so may allow certain MTB with random mutations to survive, reproduce, and create a second infection that is resistant to the antibiotic(s) originally used to treat the first infection.
A wild-type strain of M. tuberculosis does not have any gene mutations that confer antibiotic resistance, making it susceptible to all standard classes of TB antibiotics. Other strains of MTB, including MDR and XDR TB strains, have mutations in the form of single-nucleotide polymorphisms (SNPs) which lead to resistance to antibiotics. Once a bacterium is resistant to a particular antibiotic, that antibiotic is no longer effective in killing the bacteria or curing the infection.
A SNP (pronounced “snip”) is a single base pair substitution that can be observed when comparing similar DNA sequences of the same gene—either between organisms, strains, or homologous chromosomes (Figure 1a). A SNP can lead to a synonymous polymorphism or a nonsynonymous polymorphism. Synonymous polymorphisms (also known as ‘silent mutations’) do not lead to an amino acid change in the translated protein sequence, since multiple codons can code for the same amino acid (Figure 1b). Nonsynonymous polymorphisms, however, lead to a change in the protein sequence and can either cause a missense mutation, which results in a different amino acid in the protein sequence or a nonsense mutation that results in an early stop codon (Figure 1b).
Fig. 1 (a) An example of a single-nucleotide polymorphism, (b) Possible consequences of SNPs
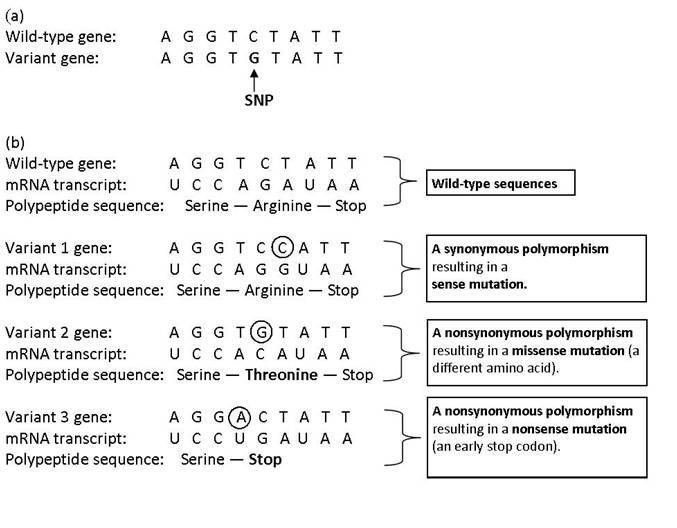
Recall that the order of amino acids in a polypeptide determines the polypeptide’s three-dimensional shape and, consequently, the protein’s structure and function.
Antibiotics often target proteins at particular binding sites that disrupt the function of the protein. Because the protein no longer works, the bacterium can’t carry out its normal functions and it will perish. For example, rifampin, an antibiotic against TB, binds to and inhibits a subunit of MTB’s RNA polymerase. If the cell’s RNA polymerase doesn’t work, what are the consequences? Genes can no longer be transcribed into proteins, and no proteins means no functional cellular machinery!
Bacteria that are resistant to rifampin can have a mutation in their rpoB gene which alters the site of where rifampin binds to RNA polymerase. Therefore, rifampin isn’t able to bind to the polymerase, the polymerase continues to work, and the bacteria live!
In this activity, you will be given the DNA sequence of the rpoB (beta subunit of RNA polymerase) gene from a wild-type Mycobacterium tuberculosis and the rpoB gene sequence of a variant strain of MTB. Your job is to identify any SNPs in the variant gene sequence, determine the amino acid sequences of both the wild-type and variant alleles and whether the SNP is a synonymous or nonsynonymous polymorphism. You will then need to critically evaluate what effect the SNP may have on conferring antibiotic resistance of the variant strain of M. tuberculosis.
Copyright © 2013 Michael Strong and Jessica Taylor